Methylation-Based Biological Age and Breast Cancer Risk
Jacob K. Kresovich; Zongli Xu; Katie M. O’Brien; Clarice R. Weinberg; Dale P. Sandler; Jack A. Taylor
J Natl Cancer Inst. 2019;111(10):1051-1058
Abstract AND INTRODUCTION
Abstract
Background: Age is one of the strongest predictors of cancer, chronic disease, and mortality, but biological responses to aging differ among people. Epigenetic DNA modifications jadwal slot Gacor hari ini have been used to estimate “biological age,” which may be a useful predictor of disease risk. We tested this hypothesis for breast cancer.
Methods: Using a case-cohort approach, we measured baseline blood DNA methylation of 2764 women enrolled in the Sister Study, 1566 of whom subsequently developed breast cancer after an average of 6 years. Using three previously established methylation-based “clocks” (Hannum, Horvath, and Levine), we defined biological age acceleration for each woman by comparing her estimated biological age with her chronological age. Hazard ratios and 95% confidence intervals for breast cancer risk were estimated using Cox regression models. All statistical tests were two-sided.
Results: Each of the three clocks showed that biological age acceleration was statistically significantly associated with increased risk of developing breast cancer (5-year age acceleration, Hannum’s clock: hazard ratio [HR] = 1.10, 95% confidence interval [CI] = 1.00 to 1.21, P = .04; Horvath’s clock: HR = 1.08, 95% CI = 1.00 to 1.17, P = .04; Levine’s clock: HR = 1.15, 95% CI = 1.07 to 1.23, P < .001). For Levine’s clock, each 5-year acceleration in biological age corresponded with a 15% increase in breast cancer risk. Although biological age may accelerate with menopausal transition, age acceleration in premenopausal women independently predicted breast cancer. Case-only analysis suggested that, among women who develop breast cancer, increased age acceleration is associated with invasive cancer (odds ratio for invasive = 1.09, 95% CI = 0.98 to 1.22, P = .10).
Conclusions: DNA methylation-based measures of biological age may be important predictors of breast cancer risk. Introduction
Chronological age is the dominant risk factor for chronic disease, cancer, and death.[1,2] Although age correlates with, and presumably acts through, the accumulation of biological changes, individuals undergo these changes at different rates.[3] Age therefore serves as an imperfect surrogate for daftar situs judi slot online gampang menang time-related biological changes. Increasing age correlates with a variety of molecular changes, including telomere shortening, differential gene expression, and mitochondrial decline,[4–6] which have in turn been investigated as potential biomarkers for the prediction of disease onset and mortality.[7–11] Among such age-associated biomarkers, those based on epigenetic modifications to DNA appear particularly promising.[12] Age-related changes in DNA methylation (DNAm) may reflect long-term changes in transcriptional regulation,[13] are widespread across the genome,[14–16] and can be used as “clocks” that are strongly correlated with chronological age.[17]
Several epigenetic clocks have been proposed,[18,19] including Hannum’s clock,[20] Horvath’s multi-tissue clock,[21] and Levine’s phenotypic age (“PhenoAge”) clock.[22] The Hannum and Horvath clocks were developed by regressing chronological age on individual CpG sites using supervised machine learning algorithms to slot gampang jackpot 2022 select the most informative set to predict chronological age.[20,21] The Horvath clock was developed using DNAm from multiple tissues whereas the Hannum clock was developed using only blood DNAm. Although the Hannum and Horvath clocks were designed solely to estimate chronological age, the Levine clock uses blood DNAm to estimate a previously developed biological age metric called PhenoAge.[22] The PhenoAge metric, based on the combination of chronological age and selected blood parameters (eg. albumin, creatinine, white blood cell count, proportion of lymphocytes, and others), was designed as a predictor of age-associated mortality. The Levine clock and its PhenoAge metric have yet to be widely examined in independent studies.
Despite differences in construction, all three epigenetic clocks are correlated with chronological age. Individuals whose clock-based estimates of age (DNAm age) are greater than their chronological age are said to have “age acceleration” that may be predictive of increased risk of morbidity and mortality.[23–26] Whether cancer risk is increased in people with situs slot online terbaik dan terpercaya 2022 age acceleration remains to be explored. Breast cancer incidence increases rapidly with age,[27] and we here report results of a large, nationwide, prospective study of methylation-based age acceleration and breast cancer risk.
Methods
Study Population
The Sister Study is a prospective cohort study of 50 884 women recruited from the United States and Puerto Rico between 2003 and 2009.[28] To be eligible, women had to be age 35–75 years, could not have had breast cancer themselves, and must have had a biological sister who was diagnosed with breast cancer. Women are re-contacted annually for information on breast cancer; the response rate for annual updates is approximately 95%. Women reporting incident breast cancer are contacted 6 months following diagnosis and asked to provide authorization to retrieve medical records. Among women for whom we obtained pathology reports, the positive predictive value of a self-reported breast cancer was 99.4%.[29] The final sample size of our case-cohort analysis included a random sample of 1289 non-Hispanic white women (91 of whom were diagnosed with in situ or invasive breast cancer before October 2016; data release 6.0) drawn from the full cohort. Cases included 1475 non-Hispanic white women diagnosed with incident in situ or invasive breast cancer during the time between enrollment and when the case- cohort was sampled (July 2014).[30] Therefore, a total of 2764 women were included in the baseline blood DNAm analysis, of whom 1566 (1475 + 91) developed incident breast cancer. Pathology reports were obtained for 1538 (98%) of the women diagnosed with breast cancer.
Informed consent and blood samples were obtained during a home visit. The study was approved by the institutional review boards of the National Institute of Environmental Health Sciences and the Copernicus Group.
Genomic DNA Processing and DNAm Age Estimation
DNA processing procedures have been previous reported.[31] Briefly, genomic DNA was extracted from aliquots of whole blood drawn using an automated system (Autopure LS, Gentra Systems) in the NIEHS Molecular Genetics Core Facility or using DNAQuik at BioServe Biotechnologies LTD (Beltsville, MD). One microgram of DNA was bisulfite-converted in 96-well plates using the EZ DNA Methylation Kit (Zymo Research, Orange County, CA). Methylation analysis was carried out at the NIH Center for Inherited Disease Research (Baltimore, MD). Samples were tested for completion of bisulfite conversion, and converted DNA was analyzed using Illumina’s Infinium HumanMethylation450 BeadChip following the manufacturer’s protocol. The arrays were analyzed with high throughput robotics to minimize batch effects.
Methylation data preprocessing and quality control were completed using the ENmix R software package.[32] This included reducing background noise with the ENmix method; correcting fluorescent dye-bias using the RELIC method;[33] quantile normalization to make overall array fluorescence intensity distribution comparable between arrays; and reducing Infinium I and II probe design bias using the RCP method. [34] Sister Study data can be requested via https://sisterstudy.niehs.nih.gov/English/coll-data.htm. Using the processed DNAm data, DNAm age was calculated using R code provided by the clock developers.[20–22]
Statistical Analysis
To capture epigenetic age effects that are independent of chronological age, we examined associations with epigenetic age acceleration. For each of the three epigenetic clocks, we defined epigenetic age acceleration by regressing DNAm age on chronologic age and calculating the difference between the observed DNAm age and the fitted DNAm age. We calibrated the fitted DNAm age both for the whole analytic sample and restricting to the random subcohort; we did not observe meaningful differences using the two approaches; therefore, we used DNAm age fit to the full dataset. We estimated age acceleration with and without adjustment for blood cell composition (BCC) because it can change with age.[23] BCC was estimated using the Houseman method.[35] In analyses adjusting for BCC, we excluded B-cell proportion to avoid model overfitting; B-cell proportion was not statistically significantly associated with chronological age and had the smallest amount of variation within the study population. DNAm age and epigenetic age acceleration metric correlations with chronological age and each other were examined using Pearson correlations and corresponding two-sided P values. We excluded any participants if the absolute value of their age acceleration estimate was greater than four SDs based on the distribution of residuals, resulting in the exclusion of 10 women (including four incident breast cancer cases) from our final analyses.
To examine associations with breast cancer risk, we used a case-cohort Cox proportional hazards model to calculate hazard ratios, 95% confidence intervals, and two-sided P values;[36] chronological age was used as the primary time-scale. In the primary analysis, invasive breast cancer and ductal carcinoma in situ (DCIS) were combined to represent all breast cancer risk. In follow-up analyses, we separately considered those categories to enable separate estimation of associations with risk of DCIS and invasive breast cancer. For the invasive breast cancer analysis, DCIS diagnosis was treated as a censoring event. Similarly, for the DCIS analysis, invasive diagnosis was treated as a censoring event. We further investigated associations restricting the risk sets by menopausal status during follow-up and considering specific outcomes based on tumor estrogen receptor (ER) status; in these analyses, we treated diagnoses with other types of breast cancer as censoring events. To reduce the potential that observed associations were a result of underlying, undiagnosed breast cancer, we repeated analyses excluding the first 2 years of follow-up. Because menopausal transition has previously been reported to increase age acceleration,[37] we conducted an analysis restricted to women who were premenopausal at baseline and tested associations with overall breast cancer risk and postmenopausal breast cancer risk. Potential effect heterogeneity for postmenopausal breast cancer risk was tested using an interaction term between the age acceleration metric and menopause status at enrollment. Finally, we used a case-only approach to examine whether age acceleration was differentially associated with specific breast cancer characteristics where the calculated two-sided P values represent a statistical test of etiologic heterogeneity.[38] We adjusted for confounding by alcohol intake (drinks per week, continuous), body mass index (BMI; kg/m2, continuous), and menopause status at study enrollment (pre- vs postmenopausal), because they may influence DNAm age[37,39] and have previously been associated with breast cancer risk in this cohort.[40,41] To account for differing effects of BMI on breast cancer risk by menopause status,[42] we included an interaction term of the two variables. Adjustment for other breast cancer risk factors, including age at menarche (years), parity (total births), and age at first live birth (years) produced only negligible changes in effect estimates. We tested the proportional hazards assumption by including an interaction term between the age acceleration metric and age at the end of follow-up, and it was not violated. We performed a complete case analysis, excluding women who were missing information on alcohol intake (n = 6), BMI (n = 2), and menopausal status at enrollment (n = 1). For all analyses, a P value of less than .05 was considered statistically significant and all tests were two-sided. Analyses were conducted using Stata version 14.2 (College Station, TX).
Results
Sample Population
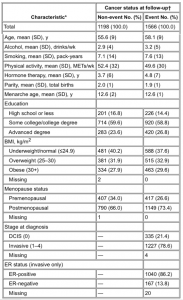
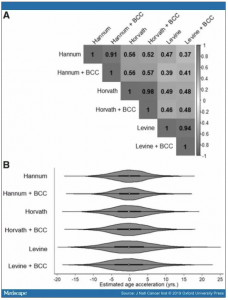
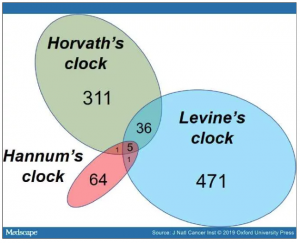
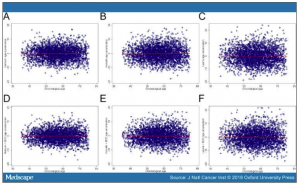
The characteristics of the sample population at enrollment are described in . As expected, all three epigenetic clocks were strongly correlated with chronological age (Hannum: Pearson r = 0.88, P < .001; Horvath: Pearson r = 0.87, P < .001; Levine: Pearson r = 0.83, P < .001) (Figure 1, A–C) . The Hannum and Horvath clocks share six CpG sites and the three clocks had five common CpGs (Supplementary Figure 1, available online). By construction, all six age acceleration metrics were independent of chronological age (all Pearson r = 0.0, all P > .7) (Supplementary Figure 2, available online). There was moderate correlation among the six age acceleration metrics, with the Horvath clock showing the smallest change following BCC adjustment (Pearson r = 0.98) (Figure 2A). The six age acceleration metrics had median values near zero with a majority of the values falling between −5 and 5 years (Figure 2B).
Table 1. Participant characteristics at study enrollment. See Table 1.
*Women with missing continuous covariate information: alcohol, 5 non-cases, 1 case; smoking, 4 non-cases, 6 cases; physical activity, 11 non- cases, 11 cases; hormone therapy, 5 non-cases, 4 cases; parity, 2 non-cases; menarche age, 1 non-case, 2 cases. BMI = body mass index; DCIS = ductal carcinoma in situ; ER = estrogen receptor; METs = metabolic equivalent of tasks.
†Cancer status defined as incident DCIS or invasive breast cancer.
Figure 1. See Figure 1.
Association between chronological age and the three epigenetic clocks. Scatterplots and Pearson correlation coefficients for chronological age and DNA methylation (DNAm) age estimated by the Hannum (A), Horvath (B), and Levine clocks (C) among non-Hispanic white women in the Sister Study.
Figure 2. See Figure 2.
Age acceleration correlations and distributions. Pearson correlation matrix of the epigenetic age acceleration metrics (A) and violin plots of the distributions (B). “Clocks” + blood cell composition (BCC) are age acceleration metrics adjusted for BCC. S
Supplementary Figure 1. See Supplementary Figure 1.
Venn diagram depicting the shared and unique number of CpG loci for each of the three epigenetic clocks. Five CpG sites were common to all three clocks.
Supplementary Figure 2. Age acceleration metric associations with chronological age. See Supplementary Figure 2.
Scatterplots of the unadjusted and BCC-adjusted age acceleration metrics by chronological age for the Hannum (A, D), Horvath (B, E) and Levine (C, F) epigenetic clocks.
Epigenetic Age Acceleration and Breast Cancer Risk
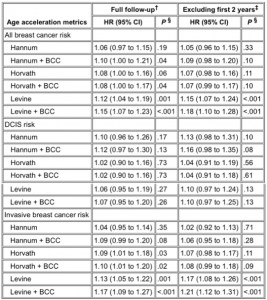
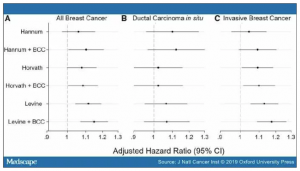
After accounting for BMI, menopause status, and alcohol intake at enrollment, BCC-adjusted age acceleration metrics by all three clocks were statistically significantly associated with breast cancer risk (Hannum + BCC: hazard ratio [HR] = 1.10, 95% confidence interval [CI] = 1.00 to 1.21, P = .04; Horvath + BCC: HR = 1.08, 95% CI = 1.00 to 1.17, P = .04; Levine + BCC: HR = 1.15, 95% CI = 1.07 to 1.23, P < .001) (; Figure 3A). After parsing cancers into DCIS and invasive breast cancer, we found that none of the age acceleration metrics was statistically significantly associated with DCIS (Figure 3B). For invasive cancers, acceleration based on both the Horvath and Levine clocks was statistically significant (Horvath + BCC: HR = 1.10, 95% CI = 1.01 to 1.20, P = .02, Levine + BCC: HR = 1.17, 95% CI = 1.09 to 1.27, P < .001) (Figure 3C).
Table 2. Epigenetic age acceleration metrics (per 5-year increase) and breast cancer risk using the full sample and excluding first 2 years of follow-up* See Table 2.
*Models adjusted for BMI, menopause status, alcohol intake at enrollment, and a BMI by menopausal status interaction term. BCC = blood cell composition; BMI = body mass index; CI = confidence interval; DCIS = ductal carcinoma in situ; HR = hazard ratio.
†Full follow-up counts: all breast cancer risk: 2754 participants, 1564 events; DCIS: 1600 participants, 334 events; invasive: 2430 participants, 1226 events.
‡After 2-year exclusion counts: all breast cancer risk: 2378 participants, 1199 events; DCIS: 1434 participants, 255 events; invasive: 2119 participants, 940 events.
§Two-sided P values calculated using Cox regression hazard models.
Figure 3. See Figure 3.
Adjusted hazard ratios (and 95% confidence intervals [CI]) for the epigenetic age acceleration metrics (per 5-year increase). Associations shown for all breast cancer risk (A) and stratified by ductal carcinoma in situ risk (B) and invasive breast cancer risk (C). “Clocks” + blood cell composition (BCC) are age acceleration metrics adjusted for BCC.
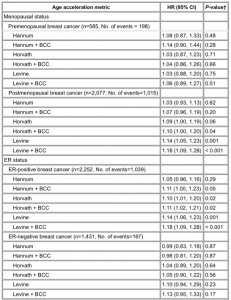
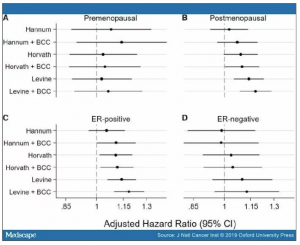
Stratification of invasive breast cancers by menopausal status at diagnosis revealed that, although there were no statistically significant associations with premenopausal breast cancer (Figure 4A; , available online), both the Horvath and Levine age acceleration metrics were statistically significantly associated with postmenopausal invasive breast cancer (Horvath + BCC: HR = 1.10, 95% CI = 1.00 to 1.20, P = .04; Levine + BCC: HR = 1.18, 95% CI = 1.09 to 1.28, P < .001) (Figure 4B). Stratification of all invasive tumors by ER status showed ER-positive invasive breast cancer risk was predicted by the Horvath and the Levine clocks (Horvath + BCC: HR = 1.11, 95% CI = 1.02 to 1.21, P = .02; Levine + BCC: HR = 1.18, 95% CI = 1.09 to 1.28, P < .001) (Figure 4C). Although none of the three clocks was statistically significantly associated with ER-negative breast cancer, the Levine metrics showed suggestive positive associations (Figure 4D).
Supplementary Table 1. Epigenetic acceleration metrics (per 5-year increase) and breast cancer risk associations for stratified by menopausal status and ER-status at diagnosis. See Supplementary Table 1.
*Models adjusted for body mass index and alcohol intake at enrollment. ER = estrogen receptor; BCC= blood cell composition; CI = confidence interval; HR = hazard ratio. † Two-sided P-values calculated using Cox regression hazard models.
Figure 4. See Figure 4.
Adjusted hazard ratios (and 95% confidence intervals [CI]) for epigenetic age acceleration metrics (per 5-year increase) for invasive breast cancer risk. Associations stratified by menopausal status at diagnosis (A and B) and tumor estrogen receptor (ER) status (C and D). “Clocks” + blood cell composition (BCC) are age acceleration metrics adjusted for BCC.
Epigenetic Age Acceleration and Time to Diagnosis
To examine the possibility that epigenetic clocks were affected by clinically occult cancer, we reanalyzed our data excluding the first 2 years of follow-up. Compared to the full follow-up, hazard ratios for all breast cancer were similar for both the Hannum and Horvath age acceleration metrics (). However, the Levine metrics were increased (Levine + BCC, before exclusion: HR = 1.15, 95% CI = 1.07 to 1.23, P < .001; after exclusion: HR = 1.18, 95% CI = 1.10 to 1.28, P < .001). In a stratified analysis, the 2-year exclusion strengthened DCIS hazard ratios for all three clocks but, for invasive breast cancer, slightly attenuated the Hannum and Horvath hazard ratios. Notably, the 2-year exclusion resulted in increased hazard ratios for invasive breast cancer using the Levine clock.
Table 2. Epigenetic age acceleration metrics (per 5-year increase) and breast cancer risk using the full sample and excluding first 2 years of follow-up* See Table 2.
*Models adjusted for BMI, menopause status, alcohol intake at enrollment, and a BMI by menopausal status interaction term. BCC = blood cell composition; BMI = body mass index; CI = confidence interval; DCIS = ductal carcinoma in situ; HR = hazard ratio.
†Full follow-up counts: all breast cancer risk: 2754 participants, 1564 events; DCIS: 1600 participants, 334 events; invasive: 2430 participants, 1226 events.
‡After 2-year exclusion counts: all breast cancer risk: 2378 participants, 1199 events; DCIS: 1434 participants, 255 events; invasive: 2119 participants, 940 events.
§Two-sided P values calculated using Cox regression hazard models.
Menopausal Transition, Epigenetic Age, and Breast Cancer Risk
We also explored whether our observed associations were affected by the menopausal transition. Although the Horvath clock may accelerate following menopause,[37] it is unclear whether women who have age acceleration before menopause have higher breast cancer risk. When we restricted our analysis to women who were premenopausal at baseline, we found that the BCC-adjusted Levine clock acceleration remained positively associated with breast cancer risk (HR = 1.18, 95% CI = 1.02 to 1.36, P = .02). Premenopausal women who transitioned through menopause during follow-up also showed increased postmenopausal breast cancer risk (HR = 1.42, 95% CI = 1.09 to 1.85, P = .009), but there was limited statistical evidence that risk from age acceleration differed by pre- and postmenopausal status at baseline (P interaction = .12). Although we did not detect associations with premenopausal breast cancer, these data suggest that age acceleration predating menopausal transition is an independent risk factor for breast cancer.
Epigenetic Age Acceleration as a Risk Factor for Breast Cancer Subtypes
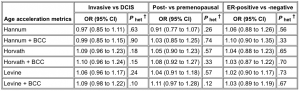
Given the possibly differential associations by invasiveness and menopausal status, we used a case-only analysis to explicitly examine effect heterogeneity. Based on the BCC-adjusted Levine metric of age acceleration, although not statistically significant, there was some evidence of case-case differences for invasive cancer vs DCIS (odds ratio for invasive = 1.09, 95% CI = 0.98 to 1.22, P = .10) (). There was no evidence of effect heterogeneity by menopause or tumor ER status.
Table 3. Case-only analysis for heterogeneity of accelerated aging effects* See Table 3.
*Models adjusted for BMI, menopause status, alcohol intake at enrollment, and a BMI by menopausal status interaction term. BCC = blood cell composition; BMI = body mass index; CI = confidence interval; DCIS = ductal carcinoma in situ; ER = estrogen receptor; OR = odds ratio.
†P het is a two-sided P value for a test for heterogeneity comparing odds of invasive vs DCIS, post- vs premenopausal, or ER-positive vs – negative breast cancer.
TABLE 1 – Click to enlarge
FIGURE 1 – Click to enlarge
FIGURE 2 – Click to enlarge
SUPPLEMENTARY FIGURE 1 – Click to enlarge
SUPPLEMENTARY FIGURE 2 – Click to enlarge
TABLE 2 – Click to enlarge
FIGURE 3 – Click to enlarge
SUPPLEMENTARY TABLE 1 – Click to enlarge
FIGURE 4 – Click to enlarge
TABLE 4 – Click to enlarge
Discussion
Using a nationwide, prospective cohort of women who were cancer-free at enrollment, we found that women with biological age estimates greater than their chronological age were at increased risk of developing breast cancer. Three different biological age estimators, all derived from DNAm, showed statistically significant associations with risk. The Levine clock had the strongest effect estimate, with each 5-year increase of biological age above chronological age resulting in a 15% increase in breast cancer risk. Separate analysis of DCIS and invasive cancers showed that age acceleration was most strongly associated with invasive cancer. Although effect sizes using Levine’s clock were similar for ER-negative and ER-positive cancers, the sample size for ER-negative cancers was smaller and associations were only statistically significant for ER-positive disease. Additional analyses excluding the first 2 years of follow-up did not diminish the effects, suggesting that results are not due to clinically occult disease and that increased age acceleration exists years before cancer development.
Chronological age is the strongest risk factor for many adverse health events, including cancer, cardiovascular, and neurodegenerative diseases, and serves as a marker for the diverse biological changes that occur over the life course.[43–45] Although single biological measures that correlate with age (eg. telomere length) can be measured directly and used for risk prediction,[5–7] the associations of these measures with disease have been inconsistent across studies.[46–49] Metrics combining multiple biological measures can have stronger correlations with chronological age and are increasingly under study as potential predictors of morbidity and mortality.[8–11] Risk markers developed using age- associated CpGs may be the most promising because they reflect, in part, shifts in transcriptional programs and possibly cumulative effects of exposures over time. These sites are often enriched at polycomb complex binding sites located in promoter regions of genes that control cell development and signaling pathways.[14–16] To the degree that DNAm is a long-term marker of transcriptional control, it potentially offers a stable biological measure for disease risk prediction. Because age-associated CpG sites are differentially methylated in tumors,[14, 50–54] including breast cancer,[55] methylation at these sites may be involved in carcinogenic processes.
Using different sets of CpGs, Hannum and Horvath’s epigenetic clocks were developed to simply predict chronological age. In contrast, the Levine clock uses CpG methylation to predict a previously proposed metric of age-related mortality, PhenoAge, that is strongly correlated with chronological age. These three clocks have only a few CpGs in common, and their resulting estimates of age acceleration are moderately correlated with one another. The CpGs that are common to all three clocks are found in the promoter regions of P2RX6, SCGN, EDARADD, IPO8, and NHLRC1; methylation of these genes has not previously been associated with breast cancer carcinogenesis. Importantly, DNAm age estimates from the three clocks do not perfectly correlate with chronological age. The divergence between a clock-based estimate of DNAm age and chronological age gives rise to the hypothesis that such differences reflect biological variation in aging; people with a DNAm age older than their chronological age would be predicted to be at increased risk for age-related disease and mortality. This hypothesis has found some support in studies using mortality endpoints.[23–26]
We find associations with breast cancer risk for all three clocks, with the largest effect sizes for the Levine clock. Although a few prior studies have examined clock associations with cancer risk, none examined risk associated with the Levine clock and all have used smaller sample populations. Among the Women’s Health Initiative, compared with cancer-free controls, the Horvath clock was accelerated for 43 women who developed lung cancer.[56] Using the Normative Aging Study, including 132 elderly men with mostly incident prostate or skin cancers, the Hannum clock was associated with both all-cancer incidence and mortality.[57] An initial study conducted within the European Prospective Investigation into Cancer and Nutrition, using 87 male colorectal cancer cases and 233 breast cancer cases, found some associations with colorectal cancer but not for breast cancer, whereas a subsequent study of 451 breast cancer cases found evidence of an association for the Horvath clock.[58,59] Using prospectively collected data, the current analysis offers the most comprehensive examination of epigenetic clocks and cancer risk to date. Our large sample afforded us the ability to examine age acceleration effects separately by tumor invasiveness, ER status, and menopausal status.
Although we find evidence that age acceleration is associated with increased breast cancer risk, our study has limitations. First, our findings on DNAm age do not provide direct mechanistic insights into how these age-related changes influence breast cancer development. Second, our study population may not reflect the general population of women at risk of breast cancer. By design, all the women enrolled in the Sister Study have a family history of breast cancer. Although age acceleration appears to be stable in adults[22] and shows some evidence of heritability[21,22] and genetic control,[60] there is no evidence to date that age acceleration is associated with family history or genetic risk of breast cancer. This hypothesis remains to be examined in more detail. Our study also has certain strengths, notably, unlike previous studies of epigenetic clocks and breast cancer risk, we accounted for factors that could bias our effect estimates, including alcohol intake and BMI, both of which are known to influence epigenetic age acceleration and breast cancer risk.[39–41,61] We also adjusted for blood cell proportions, which resulted in stronger effect estimates, perhaps reflecting subtle shifts in BCC that partially mask age acceleration associations.
In summary, we find evidence that epigenetic clocks capture age-related biological variability, which may be used to improve breast cancer risk prediction. A natural extension of these concepts may lead to the development of epigenetic clocks designed for specific chronic diseases. Such clocks may be based on known disease risk factors and might further our ability to identify populations at increased risk who may benefit from increased screening or preventative interventions.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30.
- Benjamin EJ, Virani SS, Callaway CW, et al. Heart disease and stroke statistics-2018 update: a report from the American HeartAssociation. Circulation. 2018;137(12):e67–e492.
- Belsky DW, Caspi A, Houts R, et al. Quantification of biological aging in young adults. Proc Natl Acad Sci USA. 2015;112(30):E4104–E4110.
- Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Invest. 2013;123(3):951–957.
- Sahin E, DePinho RA. Axis of ageing: telomeres, p53 and mitochondria. Nat Rev Mol Cell Biol. 2012;13(6):397–404.
- Krishnamurthy J, Torrice C, Ramsey MR, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114(9):1299–1307.
- Blackburn EH, Epel ES, Lin J. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science. 2015;350(6265):1193–1198.
- Levine ME. Modeling the rate of senescence: can estimated biological age predict mortality more accurately than chronological age? J Gerontol A Biol Sci Med Sci. 2013;68(6):667–674.
- Holly AC, Melzer D, Pilling LC, et al. Towards a gene expression biomarker set for human biological age. Aging Cell. 2013;12(2):324– 326.
- Peters MJ, Joehanes R, Pilling LC, et al. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. (https://www.nature.com/articles/ncomms9570#article-info).
- Hertel J, Friedrich N, Wittfeld K, et al. Measuring biological age via metabonomics: the metabolic age score. J Proteome Res. 2016;15(2):400–410.
- Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine. 2017;21:29–36. (https://www.sciencedirect.com/science/article/pii/S2352396417301421?via%3Dihub).
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(suppl):245–254.
- Xu Z, Taylor JA. Genome-wide age-related DNA methylation changes in blood and other tissues relate to histone modification, expression and cancer. Carcinogenesis. 2014;35(2):356–364.
- Christensen BC, Houseman EA, Marsit CJ, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602.
- Teschendorff AE, Menon U, Gentry-Maharaj A, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20(4):440–446.
- Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–384.
- Bocklandt S, Lin W, Sehl ME, et al. Epigenetic predictor of age. PLoS One. 2011;6(6):e14821.
- Weidner CI, Lin Q, Koch CM, et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014;15(2):R24.
- Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367.
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115.
- Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573–591.
- Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death.Aging (Albany NY). 2016;8(9):1844–1865.
- Marioni RE, Shah S, McRae AF, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25..(https://genomebiology.biomedcentral.com/articles/10.1186/s13059-015-0584-6).
- Christiansen L, Lenart A, Tan Q, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell.2016;15(1):149–154.
- Perna L, Zhang Y, Mons U, et al. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German casecohort. Clin Epigenetics. 2016;8:64. (https://clinicalepigeneticsjournal.biomedcentral.com/articles/10.1186/s13148-016-0228-z).
- American Cancer Society. Breast Cancer Facts & Figures 2017–2018. Atlanta, GA: American Cancer Society, Inc; 2017.
- Sandler DP, Hodgson ME, Deming-Halverson SL, et al. The sister study cohort: baseline methods and participant characteristics. Environ Health Perspect. 2017;125(12):127003.
- D’Aloisio AA, Nichols HB, Hodgson ME, et al. Validity of self-reported breast cancer characteristics in a nationwide cohort of women with a family history of breast cancer. BMC Cancer. 2017;17(1):692.
- Prentice RL. A case-cohort design for epidemiologic cohort studies and disease prevention trials. Biometrika. 1986;73(1):1–11.
- O’Brien KM, Sandler DP, Xu Z, et al. Vitamin D, DNA methylation, and breast cancer. Breast Cancer Res. 2018;20(1):70.
- Xu Z, Niu L, Li L, et al. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44(3):e20.
- Xu Z, Langie SA, De Boever P, et al. RELIC: a novel dye-bias correction method for Illumina Methylation BeadChip. BMC Genomics. 2017;18(1):4.
- Niu L, Xu Z, Taylor JA. RCP: a novel probe design bias correction method for Illumina Methylation BeadChip. Bioinformatics. 2016;32(17):2659–2663.
- Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. (https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-13-86).
- Barlow WE, Ichikawa L, Rosner D, et al. Analysis of case-cohort designs. J Clin Epidemiol. 1999;52(12):1165–1172.
- Levine ME, Lu AT, Chen BH, et al. Menopause accelerates biological aging. Proc Natl Acad Sci USA. 2016;113(33):9327–9332.
- Begg CB, Zhang ZF. Statistical analysis of molecular epidemiology studies employing case-series. Cancer Epidemiol Biomarkers Prev. 1994;3(2):173–175.
- Quach A, Levine ME, Tanaka T, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY). 2017;9(2):419–446.
- White AJ, DeRoo LA, Weinberg CR, et al. Lifetime alcohol intake, binge drinking behaviors, and breast cancer risk. Am J Epidemiol. 2017;186(5):541–549.
- White AJ, Nichols HB, Bradshaw PT, et al. Overall and central adiposity and breast cancer risk in the Sister Study. Cancer. 2015;121(20):3700–3708.
- Schoemaker MJ, Nichols HB, Wright LB, et al. Association of body mass index and age with subsequent breast cancer risk in premenopausal women. JAMA Oncol. 2018; 4(11). (https://jamanetwork.com/journals/jamaoncology/fullarticle/2685650).
- Niccoli T, Partridge L. Ageing as a risk factor for disease. Curr Biol. 2012;22(17):R741–R752.
- Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA.2005;102(30):10604–10609.
- Risques RA, Kennedy SR. Aging and the rise of somatic cancer-associated mutations in normal tissues. PLoS Genet. 2018;14(1):e1007108.
- Needham BL, Rehkopf D, Adler N, et al. Leukocyte telomere length and mortality in the National Health and Nutrition Examination Survey, 1999–2002. Epidemiology. 2015;26(4):528–535.
- Haycock PC, Burgess S, Nounu A, et al. Association between telomere length and risk of cancer and non-neoplastic diseases: a mendelian randomization study. JAMA Oncol. 2017;3(5):636–651.
- Kim S, Sandler DP, Carswell G, et al. Telomere length in peripheral blood and breast cancer risk in a prospective case-cohort analysis: results from the sister study. Cancer Causes Control. 2011;22(7):1061–1066.
- Fitzpatrick AL, Kronmal RA, Kimura M, et al. Leukocyte telomere length and mortality in the Cardiovascular Health Study. J Gerontol A Biol Sci Med Sci. 2011;66(4):421–429.
- Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2):157–158.
- Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNAhypermethylation and heritable silencing. Nat Genet. 2007;39(2):237–242.
- Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novomethylation in cancer. Nat Genet. 2007;39(2):232–236.
- Teschendorff AE, West J, Beck S. Age-associated epigenetic drift: Implications, and a case of epigenetic thrift? Hum Mol Genet.2013;22(R1):R7–R15.
- Issa JP. Aging and epigenetic drift: a vicious cycle. J Clin Invest. 2014;124(1):24–29.
- Johnson KC, Houseman EA, King JE, et al. Normal breast tissue DNA methylation differences at regulatory elements are associated with the cancer risk factor age. Breast Cancer Res. 2017;19(1):81.
- Levine ME, Hosgood HD, Chen B, et al. DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging (Albany NY). 2015;7(9):690–700.
- Zheng Y, Joyce BT, Colicino E, et al. Blood epigenetic age may predict cancer incidence and mortality. EBioMedicine. 2016;5:68–73.
- Durso DF, Bacalini MG, Sala C, et al. Acceleration of leukocytes’ epigenetic age as an early tumor and sex-specific marker of breastand colorectal cancer. Oncotarget. 2017;8(14):23237–23245.
- Ambatipudi S, Horvath S, Perrier F, et al. DNA methylome analysis identifies accelerated epigenetic ageing associated withpostmenopausal breast cancer susceptibility. Eur J Cancer. 2017;75:299–307.
- Lu AT, Xue L, Salfati EL, et al. GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat Commun. 2018;9(1):387.
- Horvath S, Erhart W, Brosch M, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci USA. 2014;111(43):15538–15543.
Funding
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences
(Z01-ES049033, Z01-ES049032, and Z01-ES044055).
Notes
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
J Natl Cancer Inst. 2019;111(10):1051-1058. © 2019 Oxford University Press